Deoxyribonucleic acid (DNA) is a charged polymer that carries the genetic information for the development, maintenance, and reproduction of organisms. The DNA double helix, or duplex, is composed of two single strands wound around each other. Double helix stability depends on the interplay of multiple interactions: base stacking, hydrogen bonding of nitrogenous base pairs, and electrostatic repulsion of charges along the backbone. These forces stabilize the double helix for a wide range of temperatures and ionic conditions relevant to the life of cells. However, these forces can get disrupted due to local conditions, like a rise in temperature to the narrow range corresponding to the melting transition. At this transition, the DNA double helix undergoes denaturation (helix-coil transition) into single strands, resulting in changes to many of its physical properties, including viscosity (Rice & Doty, 1957). Under appropriate conditions, lowering the temperature reverses this process by renaturing, annealing the DNA.
This characteristic differentiates DNA from proteins. Protein formulations exist in a thermodynamic metastable state. Proteins are incredibly heat sensitive and at temperatures not much higher than body temperature (greater than 40°C) proteins can denature. Protein denaturation is the complete loss of higher order (quaternary, tertiary, and secondary) structure in the protein leaving only the primary structure, the amino acid sequence, intact. Unlike for DNA, protein renaturation is not as common.
Temperature has a huge impact on viscosity. Viscosity reflects the molecular behavior of your samples (size, shape, interactions, microstructure), and therefore varying temperature during viscosity measurements can be very revealing and provide information about the fluid microstructure.
Healthy lungs have a mucus layer that is composed of about 1% salt, while the physiological concentration of salt is about 200 mM. Addition of salt or excipients to a DNA solution affects the melting transition temperature, which also depends on DNA concentration, ratio of G-C to A-T base pairs, number of base pairs, and arrangement of base pairs along the strand. Using viscosity measurements to characterize the melting temperature and transition range for multiple DNA concentrations and ionic conditions will aid in better understanding DNA structure and stability, which is crucial for the survival of living organisms and to the performance and outcome of molecular biology techniques like mutation scanning, genotyping, sequence matching, PCR, and microarrays.
In our application note, CT-DNA Melting Temperature, we present temperature-sweep measurements (ramp up and ramp down in temperature) of calf-thymus DNA (CT-DNA) taken with the VROC® initium one plus. We use sigmoidal fits of plots of relative viscosity vs. temperature to obtain the melting temperature at multiple DNA concentrations. Next, we perform a van’t Hoff analysis of the rescaled melting temperature vs. concentration to predict thermodynamic parameters associated with DNA structural recovery. In addition, in our application note, Structural Recovery of DNA in Multiple Ionic Conditions, we explore the effect of added ions or excipients (HEPES, NaCl, or Tris-HCl) on the melting transition and structural recovery of DNA.
In the ramp up, there is a monotonic sigmoidal rise in the relative viscosity from 40 to 70 °C. This rise in relative viscosity is likely due to DNA melting (denaturation) caused by dissociation of the hydrogen-bonded base pairs within and at the ends of the DNA strands. More dissociated regions of DNA strands contribute to more intermolecular hydrogen bonding that facilitates crosslinking, leading to increase in viscosity. For the ramp down from 70 to 40 °C, the relative viscosity decreases as the DNA single strands renature, anneal to form double strands. However, the relative viscosity is not recovered, is higher, after one cycle. Such hysteresis indicates that the DNA is not renatured/annealed to its original extent. This hysteresis can be decreased or eliminated by increasing the rate of heating and/or lowering the rate of cooling. Hysteresis behavior of melting curves of high molecular weight DNA in low ionic strength buffer is reported in the literature (Michel, 1974).
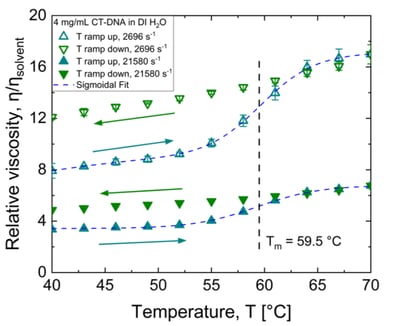 Figure 1. Viscosity of CT-DNA not recovered after one cycle of temperature ramp up and down.
Figure 1. Viscosity of CT-DNA not recovered after one cycle of temperature ramp up and down.
Extracting such parameters can be important in multiple applications to predict the sequence of DNA, like the ratio of G-C to A-T bonds, as well as its stability in different buffers and conditions.
Written by: Dr. Chrystian Ochoa, RheoSense Applications Scientist & Eden Reid, RheoSense Senior Marketing & Sales Operations



